New study shows strong interaction between genetics and nutritional supplement treatment on progression to neovascular age-related macular degeneration (AMD)
In a study published on January 8, 2018 in the Proceedings of the National Academy of Sciences (PNAS), Demetrios G. Vavvas, MD, PhD, Associate Professor of Ophthalmology at Harvard Medical School and Incumbent of the Monte J. Wallace Ophthalmology Chair in Retina at Mass. Eye and Ear, as well as investigators from University of Toronto and Stanford University, addressed a controversial topic in the field of ophthalmology – whether treatment with the Age-Related Eye Disease Study (AREDS) formulation, a combination of high-dose antioxidants and zinc, is helpful or harmful to patients based on their underlying genetics. AREDS supplementation has been widely recommended for patients with intermediate AMD since 2001.
The researchers analyzed data from 802 patients (299 of those not analyzed in prior publications)who had been treated with either the AREDS formulation or a placebo in the original 2001 study.Their analyses showed that variations in CFH and ARMS2, two genes known to influence the progression to advanced AMD, powerfully affect an individual’s response to the AREDS formulation.Approximately 40% of patients in the study had a 50% reduction in risk of progression from intermediate to advanced AMD, which was double the benefit shown in the 2001 AREDS publication.However, 15% of patients with a specific combination of genetic risk variants nearly tripled their risk of developing neovascular AMD when treated with the AREDS formulation instead of a placebo.
Similar to the original AREDS study, the investigators in this study found that AREDS formulation treatment influences disease progression only in the neovascular form of AMD, commonly referred to as wet AMD. There was no significant treatment impact on progression to geographic atrophy, the advanced dry form of AMD.
“The results of our study show that an individual’s response to the AREDS formulation is influenced by that person’s genetic makeup, and underscores the importance of using genetic testing, whenever possible, to help guide the management of patients with AMD,” said Dr. Vavvas, adding “by utilizing genetics and personalized medicine to develop more precise diagnostics and treatments, we aim to improve the overall outcomes for our patients.”
Pharmacogenomics and Age-Related Macular Degeneration
Pharmacogenomics is the study of the interaction between drugs and the human genome, in an effort to use a rational approach to maximize individual patient benefits and minimize adverse events. This potential was recognized as early as 510 BC by Pythagoras of Samos, when he noticed a connection between fava bean ingestion and hemolytic anemia in certain people. It was not until 1961 that a deficiency in G6PD was found to be responsible for favism. Around the same time, it was recognized that abnormalities in butyrylcholinesterase can result in serious adverse reactions after succinylcholine-aided anesthesia. It took another half century before we had the first FDA-approved pharmacogenetic test for cytochrome CYP2D6 and CYP2C19 alleles. The FDA requires many drugs to carry labels warning of specific gene interactions (www.fda.gov/Drugs/ScienceResearch/ucm572698.htm).
At A Glance
• Pharmacogenomics is the study of the interaction between drugs and the genome.
• More than 30 genes affect the risk for and progression of AMD.
• Used in clinical practice and in the management of patients with wet AMD, genetic information can help us better understand and treat the disease.
Genetic Influence
Age-related macular degeneration (AMD) is one of the most genetically influenced multigenic diseases found in humans, with more than 30 genes known to affect its risk and progression. 3,4 Variants of two particular genes, complement factor H (CFH) and age-related maculopathy susceptibility 2 (ARMS2), have the strongest influence on AMD development and progression. These genes have been shown not only to affect progression to the diseased state but also to affect patients’ responses to therapy.
A meta-analysis by Chen and colleagues of 13 studies involving more than 2,700 patients concluded that CFH Y402H polymorphism may play a role in patients’ response to anti-VEGF treatment for wet AMD, especially for white individuals.5 Similar results were found in the most recent and comprehensive meta-analysis, including more than 2,960 patients. Those authors reported that “individuals carrying the rs1061170/Y402H TT genotype were more likely to achieve a better treatment outcome (OR = 1.932, 95% CI = 1.125–3.317, P = .017) than those carrying the CC genotype.”6
Another meta-analysis examining the ARMS2 A69S rs10490924 risk allele in more than 2,380 patients found that patients homozygous for the low-risk allele (GG) had a higher chance of better response compared with patients with TG or TT alleles (OR 1.34; P = 0.039). However, the subgroup analysis suggested that this finding may be driven by the Asian population and may not hold true in whites.7
A more recent prospective study of 103 white patients over 4 years revealed in multivariate analysis that the ARMS2 A69S rs10490924 high-risk allele TT patients had more recurrences than the low-risk allele patients.8
There are smaller studies on single-nucleotide polymorphisms (SNPs) of VEGF-A and kinase insert domain receptor (main VEGF receptor) and response to VEGF therapy. A meta-analysis of about 440 patients revealed that only one VEGF-A SNP (rs833061) was significantly associated with treatment response,9 and a study of 377 patients investigating the major VEGF co-receptor neuropilin-1 (NRP1) suggested that patients with AA or GA NRP1 SNP rs2070296 genotype performed worse at 3 months when compared with individuals who possessed the GG genotype.10
Given the heterogeneity in study designs and resulting heterogeneous findings, it is no surprise that genetic information continues to be underused in clinical practice and overlooked in the management of patients with wet AMD.
Unlike wet AMD, for which there are efficacious treatments,11-13 there is no therapy shown to be effective for non neovascular, or dry, AMD. Only one study, the Age-Related Eye Disease Study (AREDS) Report No. 8, has shown that supplementation with high-dose vitamins and zinc in patients with advanced AMD (categories 3 and 4) can reduce progression to advanced AMD by about 25%.14 Detailed analysis reported in this study revealed that only progression to the wet form of the disease was statistically significantly affected (0.62, 95% CI = 0.43–0.90, P = .001), whereas atrophic changes showed opposing, non–statistically significant trends, for central (decreasing trend, P = .13) or noncentral (increasing trend, P not provided) geographic atrophy (GA).
Pharmacogenomics And Treatment Response
Several years after AREDS Report No. 8 was published, several authors attempted to investigate the pharmacogenomics of this supplementation, leading to controversy in the literature. Investigating a combined endpoint of progression to both neovascular and central GA stage in a post hoc fashion, Klein et al suggested that the benefit of the AREDS formulation (PreserVision; Bausch + Lomb) may be reduced in patients with high CFH risk allele.15 A subsequent post hoc study by Awh et al, considering a partial cohort of the AREDS study population, suggested that high CFH risk may actually be harmful, and that patients with the ARMS risk allele may benefit even more from use of the formulation than the average patient.16 However, as noted, that study did not include the full AREDS dataset and lacked a validation group, which is important in any retrospective analysis.
Subsequently, investigators for AREDS Report No. 38 could not replicate the interactions between genetics and response to supplementation reported by Awh et al.17 It is interesting to note, however, that AREDS Report No. 38 analyzed so many genetic subgroups and treatment variations that it resulted in small sample sizes for each subgroup and diminished overall statistical power. Thus, for each individual subgroup, the study could not identify any group with statistically significant benefit.
To increase statistical power, Awh et al subsequently analyzed only four subgroups based on CFH/ARMS2 risk alleles and replicated their original conclusions,18 still without including a validation cohort. Assel et al then further examined this controversy and could not find an interaction between supplements and genetics and progression to advanced AMD.19 It should be noted, however, that none of these studies evaluated progression only to wet AMD.
Seddon et al studied 4,124 eyes of AREDS patients and concluded that “the effectiveness of antioxidant and zinc supplementation appears to differ by genotype.”20 These authors also identified that the genetics-treatment interaction exists only for progression to the wet form of advanced AMD, and that “no significant treatment effect was observed for GA.”20 This is similar to the findings of the original AREDS Report No. 8.14
Genetics: And Important Piece Of The Puzzle
My colleagues and I investigated the interaction between genetics and supplements using wet AMD, the only statistically significantly proven event in the AREDS study, as endpoint progression.14,21 Our study included the largest cohort of AREDS patients to date, plus 103 patients from an additional cohort (N = 1,624).21 We used several accepted statistical approaches to demonstrate a strong, statistically significant dependence of treatment outcome on genetics. More important, we used a validation dataset of 299 patients that demonstrated even stronger interaction.21
These data provide further support that response to the AREDS formulation treatment differs substantially among individuals, based on genetic risk. Unlike FDA-approved drugs that must have two phase 3 randomized trials to demonstrate efficacy and safety before being allowed to go to market, there has been no placebo-controlled replication study of the efficacy of the AREDS supplements. This is because the manufacturer markets the formulation as a supplement with the disclaimer that it is not intended to treat or prevent any disease, rather than as a drug with therapeutic impact; thus, it bypasses FDA jurisdiction. Furthermore, the lack of a controlled replication-validation trial has not prevented the widespread acceptance and recommendation of the AREDS formulation treatment for patients with intermediate AMD.
If we want to better understand and treat this disease, it would behoove us to take advantage of the genetic information that we are now able to obtain. Our investigations and those of others suggest that pharmacogenomics is here to stay.
CFH and ARMS2 genetic risk determines progression to neovascular age-related macular degeneration after antioxidant and zinc supplementation
We evaluated the influence of an antioxidant and zinc nutritional supplement [the Age-Related Eye Disease Study (AREDS) formulation]on delaying or preventing progression to neovascular AMD(NV) in persons with age-related macular degeneration (AMD).AREDS subjects (n = 802) with category 3 or 4 AMD at baseline who had been treated with placebo or the AREDS formulation were evaluated for differences in the risk of progression to NVas a function of complement factor H (CFH) and age-related maculopathy susceptibility 2 (ARMS2) genotype groups. We used published genetic grouping: a two-SNP haplotype risk-calling algorithm to assess CFH, and either the single SNP rs10490924or 372_815del443ins54 to mark ARMS2 risk.
Progression risk was determined using the Cox proportional hazard model. Genetics–treatment interaction on NV risk was assessed using a multi iterative bootstrap validation analysis. We identified strong interaction of genetics with AREDS formulation treatment on the development of NV. Individuals with high CFH and no ARMS2 risk taking the AREDS formulation had increased progression toNV compared with placebo. Those with low CFH risk and highARMS2 risk had decreased progression risk. Analysis of CFH and ARMS2 genotype groups from a validation dataset reinforces this conclusion. Bootstrapping analysis confirms the presence of a genetics–treatment interaction and suggests that individual treatment response to the AREDS formulation is largely determined by genetics. The AREDS formulation modifies the risk of progression to NV based on individual genetics. Its use should be based on patient-specific genotype.
Age-related macular degeneration (AMD) is the leading cause of visual disability in the industrialized world and the third leading cause globally (1). Approximately 11 million individuals are affected with AMD in the United States, with a global prevalence of 170 million, projected to increase to 288 million by the year 2050. In 2009, the direct annual health care cost due to AMD in the US was $4.6 billion (2).AMD preferentially damages the macula, the central region of the retina (3). AMD may be classified as early, intermediate, or advanced, based on macular phenotype and visual acuity.Blindness due to AMD is typically caused by the advanced stage of the disease, which takes two principal forms.
Neovascular AMD (NV) refers to pathologic angiogenesis and its sequelae and is characterized by relatively rapid loss of central vision.Central geographic atrophy (GA) refers to localized atrophy of central macular tissue and associated structures and is characterized by a more gradual progressive loss of vision (3). The risk of developing AMD is influenced by a complex interaction of age, environment, and genetics. Genetic factors add to retinal phenotype as predictors of advanced disease (4). Polymorphisms in the complement factor H (CFH) and age-related maculopathy susceptibility 2 (ARMS2) genes have the greatest impact on the progression to advanced AMD (5–7).
The Age-Related Eye Disease Study (AREDS) concluded that the AREDS formulation, a combination of high-dose antioxidants(β-carotene, vitamin C, and vitamin E) and high-dose zinc, reduced the 5-y risk of progression from intermediate to advanced age-related macular degeneration by 25% (8). Although advanced AMD was defined in the AREDS as either NV or central GA, the demonstrated reduction in progression to advanced AMD was due to a decrease in progression to NV and not by decreased progression to central GA, even after long-term evaluation (8, 9).Multiple publications have evaluated the influence of genetic risk on the response to the AREDS formulation (10–13). These analyses are all derived from various data subsets from theAREDS, the only placebo-controlled, long-term study of the effect of nutritional therapy on AMD progression. These publications address the phenomenon of interaction, a statistical term indicating that the effect of one independent variable on a dependent variable is influenced by the level of a second independent variable.
Many of these publications have looked for a significant interaction between genetics and the AREDS formulation on AMD progression. Controversy exists based on methodology and subset of patients analyzed. In particular, Awh et al. (12) have stated that genotype groups, defined by combinations of variants in the CFH and ARMS2 genetic regions, can identify individuals who benefit greatly from treatment with the AREDS formulation, as well as those who derive no benefit, or are maybe even harmed. Chew et al. (14)found no evidence of genetic influence on response to AREDS formulation treatment.
These analyses included both central GAand NV as clinical end points. Earlier work by Chew et al. (8, 9)noted that AREDS supplements do not delay or prevent centralGA, and Seddon et al. (11) found a significant interaction between genetics and AREDS formulation treatment on NV progression but not for central GA. AREDS formulation treatment delays or prevents progression only to NV, and the inclusion of patients who progress to central GA in these analyses dilutes the data and may obscure a significant interaction.Because the subsequent AREDS2 was designed and conducted without a placebo control arm, no large dataset exists to validate either the primary or any secondary findings from the AREDS. In this study of an expanded dataset from the AREDS, we perform a validation analysis of the interaction of genetics and treatment using bootstrapping, a statistical resampling technique. We used 0.632 bootstrapping to compare the predictive accuracy of models of NV progression risk that may or may not include interaction of genotype group with AREDS formulation treatment.
By aggregate analysis of multiple (thousands) random discovery and validation sets generated through resampling of the main dataset, predictors of NV progression can be accurately identified by observing the incremental contribution to model accuracy. This well-established computational method allows powerful statistical determination of the reproducibility of prediction models (15). Validation using the bootstrapping technique can help distinguish false associations,resulting from overfitting or multiple testing, from true ones. Bootstrapping accurately identifies true determinants of clinical outcome better than analysis of a single dataset (15–17).
Materials and Methods
Subjects
Subjects were derived from the AREDS population. Study procedures have been reported previously (8). Subjects were characterized by AREDS investigators at enrollment and during half-yearly follow-ups using retinal images classified by a central reading center. This allowed determination of the time interval from study enrollment to AMD progression to either central GAor NV (8). The AREDS investigators randomized subjects to receive placebo,zinc (80 mg daily), antioxidants (β-carotene, 15 mg; vitamin C, 500 mg; and vitamin E, 400 IU), or both zinc plus antioxidants. Our analyses are restricted to subjects randomized to placebo or to zinc plus antioxidants (the “AREDS formulation”). Subjects who experienced progression events at 2 y or less from study enrollment were not considered in this analysis, since these events were unlikely a result of the assigned treatment (9). The complete phenotype data were provided through the database of Genotypes and Phenotypes (dbGAP)under an investigator agreement with one of the authors (R.K.). This work was approved by the University of Toronto Research Ethics Board. Informed consent was provided by all study subjects upon enrollment in the AREDS.
Genetic Datasets
We assembled genotyping data from three separate sample sources: (i) Targeted sequencing was performed by others on 3,340 AMD samples from the Michigan, Mayo, AREDS, Pennsylvania (MMAP) sample set. Short-read sequences were matched to the Genome Reference Consortium build 37 (GRCh37) assembly before being deposited into the NIH’s dbGaP database that has been made available through an investigator agreement(R.K.). We obtained the aligned sequences from dbGaP using the NIH’s sra toolkit(version 2.5.4). The read sequences for the CFH (chromosome 1) andARMS2 (chromosome 10) loci were processed using the Samtools package(www.htslib.org) to deduce unphased genotypes at single-nucleotide polymorphic variants in the complement factor H genomic region (rs1061170,rs3766405, and rs412852) and one SNP (rs10490924) in the age-related maculopathy sensitivity 2 region (www.htslib.org). This yielded genotypes for2,003 AREDS samples of all presenting grades and treatment groups. (ii) Genotyping data were generated from 1,390 AREDS DNA samples purchased from the Coriell Institute (13). Beckman Coulter Genomics according to Good Laboratory Practices performed genotyping at CFH and ARMS2 using bidirectional sequencing. (iii) Genotypes at CFH rs2755405 and rs412852 andARMS2 rs10490924 loci from 534 cases referenced by Chew et al. and collaborators(18, 19) were made available to our group in May 2017 from the National Institutes of Health Office of Research Integrity and Compliance.Of these samples, we eliminated duplicates and all MMAP samples obtained from subjects who were not part of the AREDS. This resulted in 1,626 samples. Of these, 802 were from subjects randomized to treatment with either placebo or the AREDS formulation, which we refer to as the “expanded” dataset. Of these 802 subjects, 299 had not been part of the prior Awh et al. (12) published analyses.This subgroup of 299 subjects is referred to as the “unique” dataset. Subject distribution between treatment and genetic groups is provided in Fig. 1. AREDSID/ID2 numbers for each study subject are included as SI Appendix, Table 1.

Marker Selection
To analyze the common genetic variability of the CFH locus, we selected rs3766405 and rs412852 to tag the two major CFH haplotypes as has been done previously (12, 13). We defined the two SNP “high-risk” haplotypes to bers3766405 CC/rs412852 CC, the average-risk haplotype to be rs3766405 CT/rs412852CT or rs3766405 CC/rs412852 CT, and the “low-risk” haplotypes to be all other combinations. The derivation of these groups has been described previously (12).We prespecified genotype groups in the manner described by Awh et al. (12).Briefly, we determined the number of AMD risk alleles at CFH and ARMS2 for each subject. Given the relative rarity of homozygous CFH low-risk alleles andARMS2 homozygous high-risk alleles, subjects homozygous for these rare all eles were grouped with subjects heterozygous for the corresponding risk alleles(12). Genotype group (GTG) 1 was composed of subjects with low/intermediate CFH and no ARMS2 risk alleles (C01A0). GTG2 subjects had high CFH and no ARMS2 risk alleles (C2A0). GTG3 subjects had low/intermediate CFH and one or two ARMS2 risk alleles (C01A12). GTG 4 subjects had high CFHand one or two ARMS2 risk alleles (C2A12). This is summarized in Table 1.
Clinical Outcome Determination
Subjects in the AREDS cohort varied based on baseline AMD status. Subjects were classified based on the severity of AMD in each eye. We restricted this analysis to subjects with category 3 or 4 AMD at baseline, which are the subgroup of subjects for whom the AREDS formulation was reported beneficial in the original AREDS analysis (9).
Progression of each subject to either NV or central GA was determined through data from within dbGaP tables pht000375.v1.p1.c and pht000376.v1.p1.c1, which provide detailed disease phenotype data for each timed study visit. Definitions for progression to NV or to central GA are documented in“AREDS dbGAP Data Tables: A User’s Guide” (20) or in published work from the AREDS retinal image reading center (21). NV was indicated by a score of 11 or 12 in the AMDSEV[R/L]E data field, while central GA by a score of 10 or 12. We have censored observations at 7 y, as has been done in most previous analyses of these data. Since fundus photographs were taken starting at year 2 (20), we also eliminated any progression events reported within the first 2 y, as these were unlikely to be the result of treatment assignment.
Statistical Analysis
The main analyses done in this paper were performed withthe expanded dataset, to maximize statistical power. We note that the selection of particular biomarkers and the composition of genotype groupings in Awhet al. (12) were based on a subset of this expanded dataset. This fact could potentially bias our results, causing us to overestimate significance. To guard against this possibility, we have replicated the main results using just the unique set, that is, those subjects who were not used in Awh et al. These validation analyses appear in Analysis Restricted to the Unique Set.Analyses of genetic and nutritional supplement effects and their interactions were done using a Cox proportional hazards model that was adjusted for the following known potential confounders: age at enrollment, sex, and smoking status. Body mass index was not used as a confounder because of a sizable number of missing records. All analysis was done using R statistical software(https://cran.r-project.org) and the rms package (biostat.mc.vanderbilt.edu/wiki/Main/Rrms). Hazard ratios (HRs) and P values were calculated using the contrast()function in the rms package to compare specific groups. All statistical code used in this paper is included as SI Appendix. This permits the reproduction of all statistical calculations by any investigator with access to AREDS phenotype and genotype datasets for subjects identified in SI Appendix, Table 1.
Bootstrap Resampling Validation of Interaction
To evaluate the presence of a genetics–treatment interaction, we used the bootstrapping technique (22,23). This technique of random sampling with replacement provides a convenient,though computationally intensive, method to make population wide statistical inferences (Fig. 1). A version called “0.632 bootstrap” is widely used to validate clinical prediction models (24–26). In our setting, the bootstrap exercise was used to assess whether the previously proposed hypothesis of genetics–treatment interaction was a spurious finding or had clinical validity (12). The bootstrap results in this paper were obtained using the R package “pec” (prediction error curves), which uses 0.632 bootstrap for predictive analysis of time-to-event data (27). The statistical models behind the methods used in the pec package utilize the inverse probability of censoring weighted estimators to deal with censored time-to-event data.
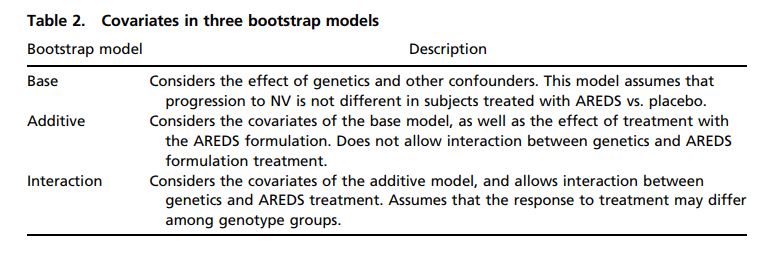
We compared event prediction accuracy in subjects using models with andwithout a genetics–treatment interaction. This approach allows validation of this interaction effect by observing whether prediction models which include theinteraction predict the outcome of subjects better than models that do not.Because the purpose of this bootstrap analysis is to test the hypothesis that agenetics–treatment interaction exists, we limited our analysis to the two genotypegroups found to be most differently affected by AREDS formulation treatment:individuals GTG2 (n = 107) and GTG3 (n = 305), for a total of 412 subjectsfrom the expanded set. In each of 100,000 iterations, 412 subjects meeting thesecriteria were randomly selected with replacement from the 412-subject set. ThreeCox models were built using these discovery sets. Study subjects not selectedbecame a paired iteration-specific bootstrap validation sample. On average, 63%of subjects would be selected randomly at least once as a discovery set, for Coxmodeling, leaving 37% for bootstrap validation. Three models were considered:(i) the base model, which considers genetics, sex, smoking, and age as covariates;(ii) the additive model, which considers the presence of AREDS formulationtreatment in addition to the base model covariates and assumes that the effectof AREDS formulation treatment is not influenced by genetics; and (iii) the interactionmodel, which considers the same covariates as the additive model butallows for the interaction of AREDS formulation treatment and genetics (Table 2).We also performed the same bootstrap model comparison on all four genotypegroups and for subjects found only within the unique set (SI Appendix, Table 1).
The predictive accuracy of each model was expressed as the concordance index (Cindex) (Fig. 2). For a pair of subjects randomly selected from a bootstrap validationset, if one subject experienced progression before the other subject, and the modelbeing evaluated correctly predicted this, that pair of subjects is considered “concordant.”The percentage of concordant pairs in each validation set with at leastone event and no event-time “ties” is the C index. One C index is generated foreach bootstrap iteration and the result is averaged over all 100,000 iterations. Thisprocedure was repeated for a number of time points within the AREDS follow-uptime range. For convenience, the C index was converted to a Somers’ Dxy measureusing the formula Dxy = 2*(C index − 0.5). Both the C index and Somers’ Dxyprovide the same information as the area under a receiver operator characteristiccurve in uncensored data, but Somers’ Dxy corrects for random guessing: It ispositive if model predictions are better than random guessing, with a maximumvalue of 1.00 indicating perfect concordance. To generate approximate pointwise95% confidence intervals, we used quantiles from a block-bootstrap approach (23),where we considered 100,000 bootstrap replications to be 1,000 realizations ofbootstrap curves, with each curve based on 100 replications.
Results
Sample Set
Data derived from purchased Coriell AREDS DNAand dbGAP MMAP sequencing and data provided by the NIH Office of Research Integrity and Compliance allowed us to identify 802 AREDS subjects (the expanded dataset) with AREDS category 3 or 4 AMD at study entry treated with either the AREDS formulation or placebo. Of these, we designate 299 subjects not used in the previous Awh analyses as the unique dataset.
Subjects receiving AREDS formulation treatment and those receiving placebo were balanced with respect to the distribution of CFH or ARMS2 risk alleles, smoking, education level, sex, and age, reflecting random AREDS treatment assignment (data not shown).